Jude Reilly
Introduction
The introduction of image search capabilities to kdb.ai has many ways that can be utilised to the user’s benefit. One way that has been shown to be a valuable use-case is the using images of CT brain scans to help doctors identify brain tumours. This can be very helpful for when doctors aren’t able to get access to specialists who can identify different tumours with ease and must use their own less specialised knowledge to attempt to provide an (albeit educated) guess as to categorise tumours. This is obviously not ideal for something as delicate as brain tumours, and so having a tool that can help with this classification is quite helpful and could be vital for helping in locations where resources might be limited.
In this vein, I have looked at another use of kdb.ai within the medical imaging field and chose to look at X-ray scans of patients’ lungs, with the goal of being able to identify if a scan of a lung shows signs of COVID-19, among other lung illnesses. While this might seem slightly outdated, as COVID is significantly more controlled than in previous years, it sets a good precedence of how kdb.ai can be used to help with similar diseases that are either newfound diseases that no one has had time to become specialised in, or just for other diseases that doctors need help identifying due to a lack of specialised knowledge.
Our data
The dataset in use for this blog was taken from Kaggle – COVID-19 Radiography Database (kaggle.com)
This dataset is comprised of different X-ray images of lungs taken from several sources, including padchest, medical schools, Github, SIRM and other datasets within Kaggle. It was collated by a research team from Qatar University and University of Dhaka in collaboration with medical doctors from Pakistan and Malaysia. It includes X-ray images for COVID-19 positive test cases along with Normal, Viral Pneumonia, and Lung Opacity (Non-COVID lung infection) images. The dataset included 10192 images of Normal lungs, 3616 images of COVID-19 lungs, 6012 images of Lung Opacity lungs, and 1345 images of Viral Pneumonia lungs.
The Neural Network
Before introducing kdb.ai, I first had to train a neural network on this data. Due to the large size of the data, I decided that training on a large subset of the data, rather than the whole dataset, would be sufficient to still successfully train the model, whilst reducing the amount of computation time needed to perform the training, as for a dataset this size, training the neural network would take a significant amount of time. This ended up being the case, as with even 70% of the data being used, the neural network took over 10 hours to train itself on the data. Also, by keeping 30% of the data as not a part of what data is used to train the neural network, this means that when it comes to actually trying out the model in conjunction with KDB.AI, the neural network is not biased by having been trained on the images that we are inserting into it to perform image search.
At this point I should note that the neural network model I chose to use for this dataset is the ResNet50 model, the same used for the aforementioned CT brain scans use case. We at Data Intellect have looked comparing different neural networks, as you can see here. From this blog, we can see that some neural networks perform well with KDB.AI better than others, but the initial analysis for this current blog was done using ResNet50, and so is the one used throughout.
So, after loading in my pre-trained neural network, I create embeddings for all the images I did not use to train the model and assign each a label based off what category it belongs to, i.e. COVID, Normal, Viral Pneumonia or Lung Opacity. I then created a DataFrame which includes the source of the image, the class it belongs to, and the embeddings of each image.
Embeddings on full display
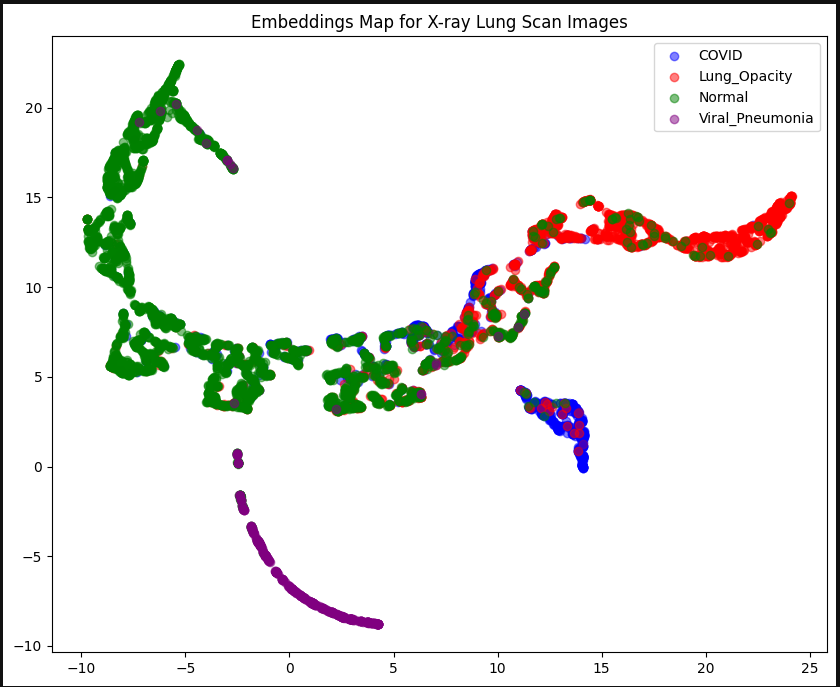
As you can see, each of the four categories are quite clustered together on the graph. There is a wide spread of Normal lungs that does crossover with other categories. This may be because there is a significantly larger amount of Normal lung images in our dataset than all the other categories. This is due to the dataset we are working with. We may have a clearer distinction between categories if we had a similar number of images for each category, but there are less images available of diseased lungs, unfortunately (purely for the purpose of data analysis!). We could also reduce the number of Normal images we use in the dataset, but for the purposes of data integrity, we shall continue with the data we have.
Introducing KDB.AI
Now KDB.AI comes into play. We now start up our instance of KDB.AI to store the embeddings. We first create the schema for our table, which is where we define our indexing and similarity metrics. Initially I chose to go with the HNSW indexing method, and Euclidean distance as my method of similarity, but we will see later how changing either of these affects the accuracy of our model.
After creating the schema, we then create our table, and then we must insert our embeddings into it. At the moment, kdb.ai requires uploading data chunk no more than ≈10MB in size at a time. Due to the large size of the dataset we are working with, this means we must split up our data into small enough chunks, and then upload these one at a time to the table.
Now that we have the data uploaded to our KDB.AI table, we can start performing image search. We will be using images that are already stored within the database as our ‘search term’, so when we search for nearest neighbours, the actual image itself will be included within our search, so when we search for the 9 nearest neighbours, we’re getting back the image itself as the nearest neighbour, plus 8 more. So, we look at the nearest 8 neighbours of our search image, and we expect that most, if not all the nearest neighbours we see will be of the same class as our original search image.
After trying some random index numbers to see what outputs were given for our nearest neighbours, I decided to try developing a function that would look at the nearest neighbours as determined by KDB.AI and assign a weighted average accuracy value based off what those nearest neighbours are.
Example 1
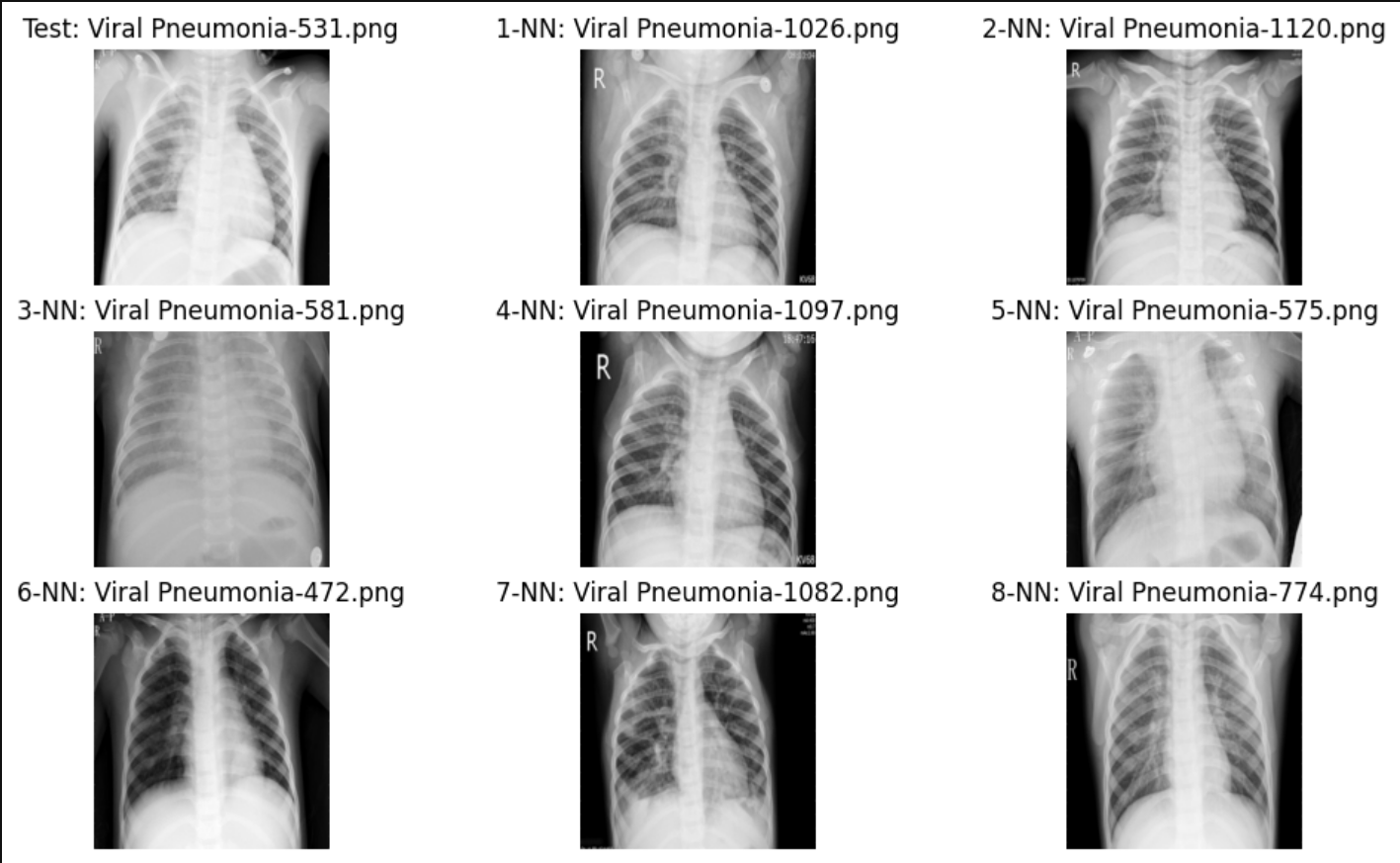
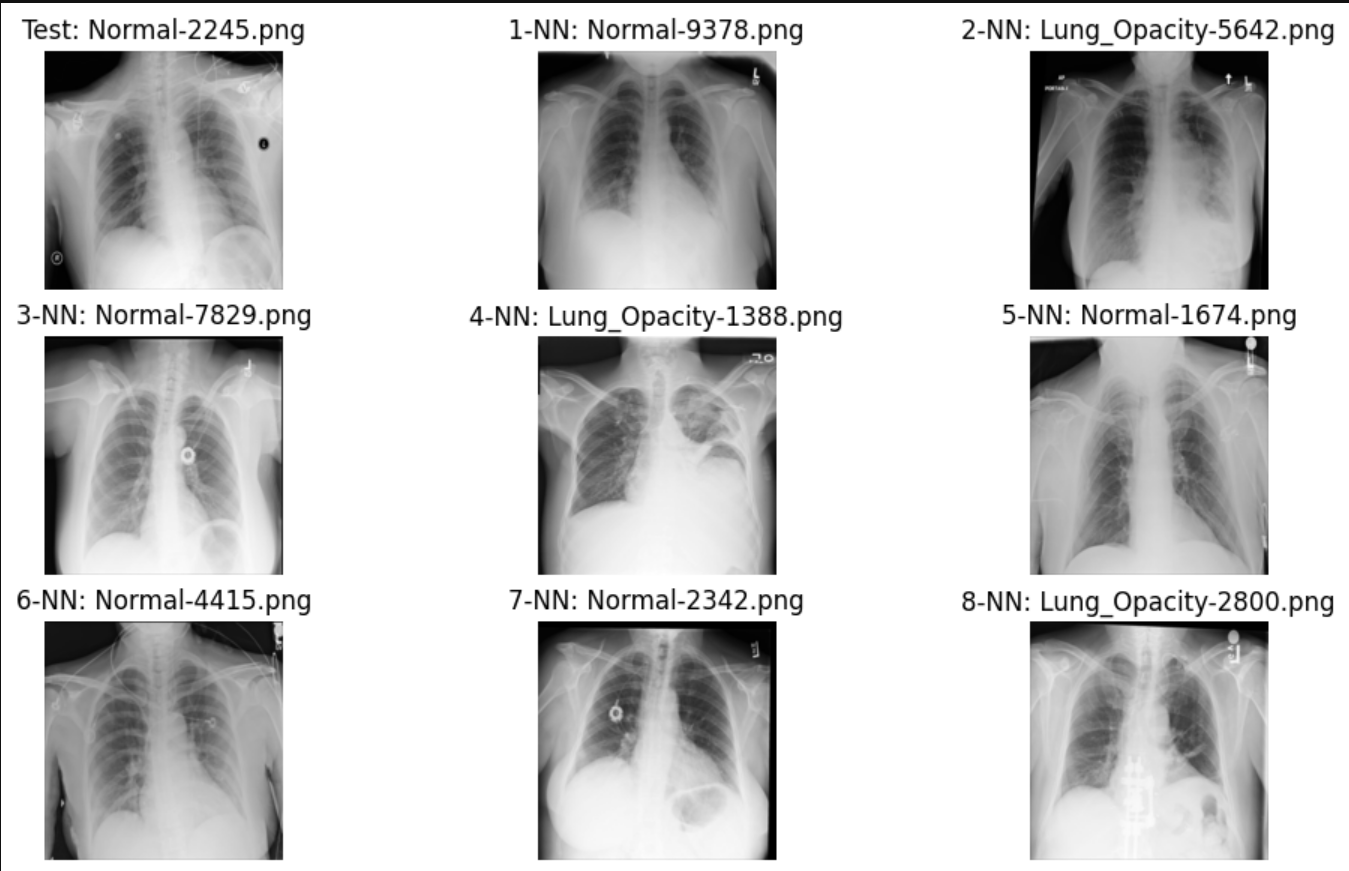
Example 2
After developing this accuracy metric, I found that this model, with this setup, had an accuracy of 76.2%. This is a fairly good level of accuracy, but now I want to look at how using different similarity metrics affects the accuracy. So, I created two other tables where the schemas have the other two similarity metrics, dot product and cosine similarity. After running the accuracy measure function on these tables, cosine similarity also had an accuracy of 76.2%, while dot product had an 80.4% accuracy. This fits well with our initial research into KDB.AI, where we found that, out of all the similarity metrics, dot product is generally the best metric to use for areas such as image/video comparisons and music. And we can see here the data shown again supports that claim.
Finally, for posterity, I decided to see how building the database using different indexing methods, and then combining them with all the similarity metrics, yields the highest accuracy.
|
|
Euclidean distance | Dot Product | Cosine Similarity |
| HNSW | 76.2% | 79.9% | 76.2% |
| Flat | 76.3% | 80.4% | 76.3% |
As we can see above, switching to a flat table structure does improve accuracy over HNSW (albeit only slightly), which again we would expect to find after our initial research. Flat file structures do indeed improve accuracy; however, their drawbacks lie with their speed and memory efficiencies. But due to the relatively small database we are working with here, and the lack of a real-time environment, those drops in speed are of little concern for us, as we just want pure accuracy with our data. And in fact, Flat indexes drop in speed comes from its retrieval, and its indexing speed is in fact faster than HNSW, so due to the fact we are working with such a relatively small dataset means that Flat ends up being overall faster than HNSW. Those of you familiar with KDB.AI may notice a noticeable absence of one indexing method – IVFPQ. This method was not included in this analysis, as IVFPQ is a method reserved almost exclusively to text, due to its inherent nature of quantizing and indexing words from documents.
So, we can see that our best combination was a Flat index structure, with a dot product similarity metric, which produced an accuracy of 80.4%. And as mentioned before, this accuracy is likely affected by the large proportion of Normal lung images vs the COVID/Lung Opacity/Viral Pneumonia images. I daresay a similar number of each type of lung scan category would lead to a fairer training of the neural network, as well as meaning our indexing with KDB.AI would be more accurate. And remember, this 80.4% accuracy value is a measure of how accurate the 8 nearest neighbours we get when performing an image search with KDB.AI is in terms of our neighbours being in the same class as our ‘search term’ image. Since the goal here would be for a professional medical practitioner to be observing these nearest neighbours, and determining what our original image should be classified as based off those neighbours, we can allow for a slight drop in accuracy, as the human observing these images would be able to use common sense, intuitiveness, and medical expertise to help bridge this gap to perform incredibly accurate classifications at, we expect, a higher rate than if they didn’t have our KDB.AI Image Search tool at their disposal.
Conclusion
So overall, we can expect that KDB.AI Image Search can used as an exceptional approach for medical diagnostics. Through the power of image recognition, healthcare professionals can expedite the identification process, distinguishing between healthy and diseases lungs with greater accuracy and efficiency. The implications of what this could lead to are profound. Had this technology been available when COVID was only burgeoning, we may have been able to classify COVID patients significantly earlier, and been able to provide treatment and containment swiftly, potentially leading to a reduced spread of the disease.
As we have also seen, this can extend to other diseases, as well. Diseases like viral pneumonia can be identified earlier, leading to timely treatment and interventions, ultimately improving outcomes and saving lives. Moreover, integration of image search technology into the medical industry could lead to streamlining workflows, reducing diagnostic errors, and alleviating burden on already pressures healthcare systems worldwide.
As we navigate the complexities of healthcare and it’s continuing further integration with technology in the 21st century, the emergence of innovations like KDB.AI highlights the potential of technology in reshaping medical diagnostics. Embracing these advancements and fostering collaboration among tech developers, healthcare providers, and policymakers is vital. The advent of KDB.AI in medical diagnostics promises a future of precision and efficiency for global healthcare – which I think we can all agree, would be a bit of a breath of fresh air for us all.
Share this: